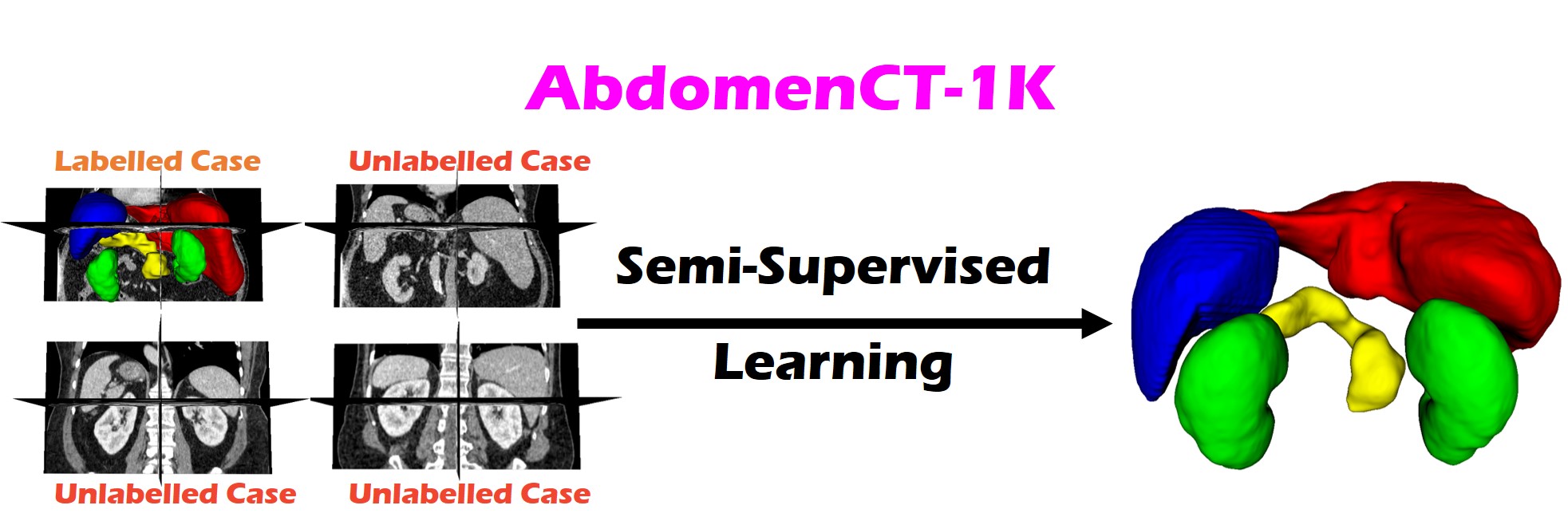
AbdomenCT-1K: Semi-supervised Learning Benchmark¶
Abdominal organ segmentation plays an important role in clinical practice, and to some extent, it seems to be a solved problem because the state-of-the-art methods have achieved inter-observer performance in several benchmark datasets [1-3]. However, it is unclear whether the excellent performance can be generalized on more diverse datasets. Moreover, to alleviate the dependency on annotations, semi-supervised learning, weakly supervised learning, and continual learning have been active research topics, but there are still no segmentation benchmarks for these tasks.
To address these limitations, we establish four benchmarks for multi-organ segmentation, including:
- AbdomenCT-1K: Fully Supervised Learning Benchmark
- AbdomenCT-1K: Semi-supervised Learning Benchmark
- AbdomenCT-1K: Weakly-supervised Learning Benchmark
- AbdomenCT-1K: Continual Learning Benchmark
Fully annotated multi-organ cases are usually expensive to be obtained while unlabelled cases are relatively easy to collect. Therefore, we set up AbdomenCT-1K: Semi-supervised Learning Benchmark for multi-organ segmentation, which focuses on learning with limited labelled cases and many unlabelled cases.
Participants are required to develop semi-supervised segmentation methods that can segment the liver, kidney, spleen, and pancreas simultaneously. This benchmark consists of 3 subtasks in consideration of the source and amount of unlabelled data. Please refer to the Dataset page for more details.
How to participate¶
- Click on the Join button.
- Download the training and testing data on the Dataset page.
- Develop your solution and make a complete submission (including a zip file of segmentation results and a short paper).
Rules¶
- All participants should register this challenge with their real names, affiliations, and affiliation E-mails. Incomplete and redundant registrations will be ignored without notice.
- For a fair comparison, participants are not allowed to use any additional data and pre-trained models.
- Participants are not allowed to register multiple teams and accounts.
Evaluation Metrics¶
¶
- Dice Similarity Coefficient (DSC)
- Normalized Surface Dice (NSD)
The implementation is available here.
Reference¶
[1] P. Bilic, P. F. Christ, E. Vorontsov, G. Chlebus, H. Chen, Q. Dou, C.-W. Fu, X. Han, P.-A. Heng, J. Hesser et al., "The liver tumor segmentation benchmark (lits)," arXiv preprint arXiv:1901.04056, 2019.¶
[2] F. Isensee, P. F. Jaeger, S. A. A. Kohl, J. Petersen, and K. H. Maier-Hein, "nnu-net: a self-confifiguring method for deep learning" based biomedical image segmentation,” Nature Methods, vol. 18, no. 2, pp. 203–211, 2021.¶
[3] N. Heller, F. Isensee, K. H. Maier-Hein, X. Hou, C. Xie, F. Li, Y. Nan, G. Mu, Z. Lin, M. Han et al., “The state of the art in kidney and kidney tumor segmentation in contrast-enhanced ct imaging: Results of the kits19 challenge,” Medical Image Analysis, vol. 67, p. 101821, 2021.¶